Transcriptional regulation of aluminum and phosphate stress-responsive genes in plants
Funded by the NSERC Discovery Grants Program.
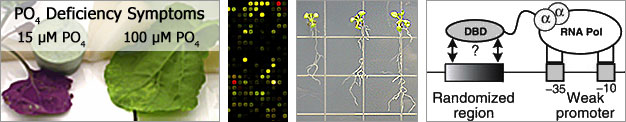
Acid soils are a major limitation to agricultural production worldwide. Factors that limit plant growth on these soils include aluminum (Al) toxicity and phosphate (Pi) deficiency. As a result, plant adaptation to acid soils requires mechanisms that confer tolerance to Al and mechanisms to enhance Pi acquisition. Preliminary analysis of gene expression experiments (transcriptome profiling of Al stress in Arabidopsis and Pi stress in canola) has identified an array of genes responsive to both stresses (co-expressed, potentially co-regulated). One striking observation was that both Al and Pi stress induced changes in expression of genes coding for many enzymes involved in carbon metabolism (glycolysis). An intriguing possibility is that increases in carbon flow through glycolysis are required to supply stress-specific metabolites not only for resistance to Al and Pi stress, but also for other abiotic stresses requiring a large allocation of carbon resources. To gain insight into plant responses to Al and Pi stress, we must identify and characterise the regulatory network(s) involved in these stress responses. The goal of the proposed research is to use current genetic, molecular, and computational tools to identify regulatory components of the Al and Pi stress response system(s) in Arabidopsis, and provide insight into the regulation of these pathways and the relationships they share. To identify components of the regulatory network(s), we will use two complementary techniques that detect interactions between DNA-binding proteins (e.g. transcription factors) and the DNA sequences to which they bind (e.g. cis-regulatory elements). The knowledge gained from this research will aid our long term goal of developing crop species better able to tolerate acid soils.